NMR Relaxation Theories and Calculations for Dynamical Biomolecules
Many macromolecules (such as carbohydrates, small RNAs, unfolded or partially-folded proteins, multidomain proteins, and intrinsically disordered systems) might be quite flexible, therefore NMR observables do not always correlate with a single conformer but instead with an ensemble of low free energy conformers that can be accessed by thermal fluctuations. Some of my projects at Rutgers University include developing NMR relaxation theories and interpreting experimental NMR quantities (such as R1/R2/NOE, RDC and J couplings) in terms of conformational ensemble and multiscale dynamics of biomolecules.
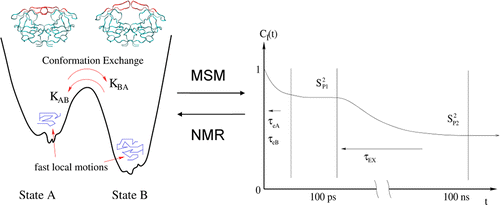
- Junchao Xia, Nan-jie Deng, and Ronald M. Levy, " NMR relaxation in proteins with fast internal motions and slow conformational exchange: model-free framework and Markov state simulations," J. Phys. Chem. B 117, 6625 (2013).
- Junchao Xia, Claudio J. Margulis, and David A. Case, " Searching and optimizing structure ensembles for complex flexible sugars," J. Am. Chem. Soc. (communication) 133, 15252 (2011).
- Junchao Xia and David A. Case, " Sucrose in aqueous solution revisited, Part 2: Adaptively biased molecular dynamics simulations and computational analysis of NMR relaxation," Biopolymers 97, 289 (2012).
- Junchao Xia and David A. Case, " Sucrose in aqueous solution revisited, Part 1: Molecular dynamics simulations and direct and indirect dipolar coupling analysis," Biopolymers 97, 276 (2012).