Structural Ensemble and Slow Dynamics of Multidomain Protein Enzymes and Signaling Proteins
Structural flexibility and slow dynamics of multidomain proteins play many roles in biological phenomena such as enzymatic functions and interaction networks responsible for cell signalling. Characterizing the interdomain motions of these proteins in wild-type and mutation is crucial to understand those mechanisms and develop corresponding control methods for diseases. For these application studies, I used various advanced sampling methods such as map-restrained Langevin dynamics, Markov state modeling and metadynamics.
- Jill J. Bouchard, Junchao Xia, David A. Case, and Jeffrey W. Peng, " Enhanced sampling of interdomain motion using map-restrained langevin dynamics and NMR: Application to Pin 1," J. Mol. Biol. 430, 2164 (2018).
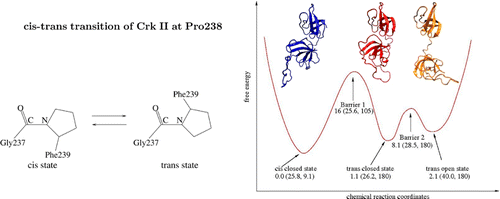
- Junchao Xia and Ronald M. Levy, " Molecular dynamics of the proline switch and its role in Crk signaling," J. Phys. Chem. B 118, 4535 (2014).
- Junchao Xia, Nan-jie Deng, and Ronald M. Levy, " NMR relaxation in proteins with fast internal motions and slow conformational exchange: model-free framework and Markov state simulations," J. Phys. Chem. B 117, 6625 (2013).