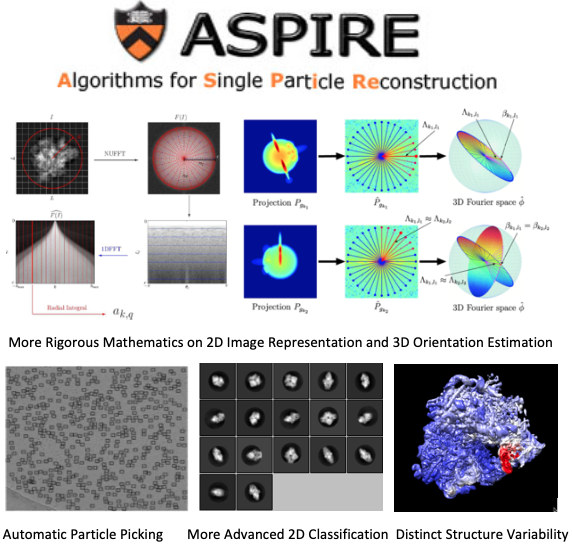
Computational CryoEM
Recent progress on computational algorithms and software is one of the major reasons leading the revolution of resolution in 3D structure determination of biomolecules using the CryoEM technique, a Nobel prize winning method projecting rapidly frozen and randomly orientated 3D particles into 2D noisy images on micrographs and reconstructing 3D density maps in atomic resolution through computer software. As the latest project with Professor Amit Singer at Princeton University, I am developing mathematical algorithms and the ASPIRE Python package for reconstructing 3D structures of biomolecules from corresponding 2D images with very low signal-noise ratio.